Gallery#
Dissociation of oxygen on Pt(100)#

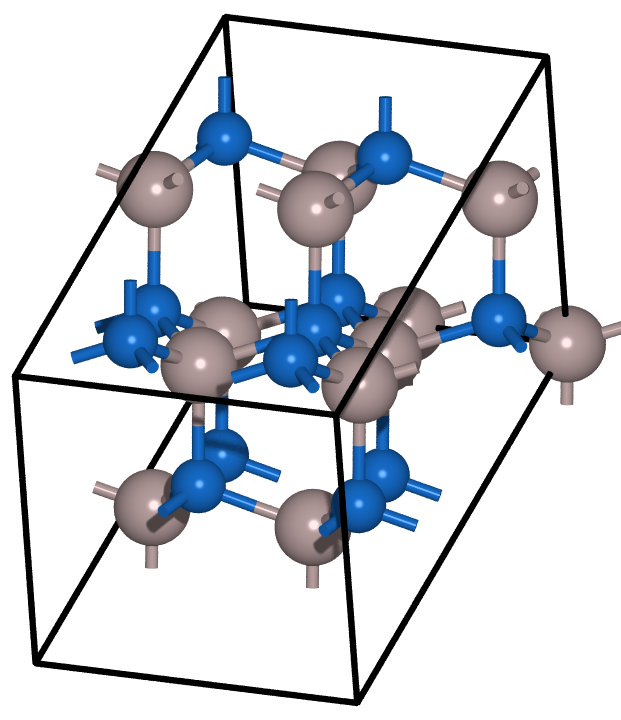
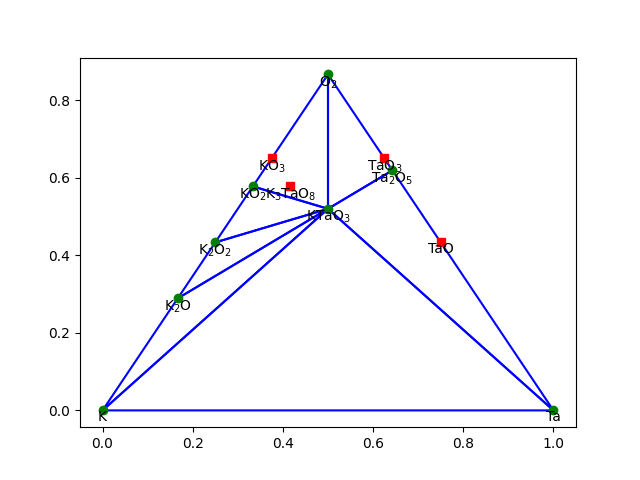
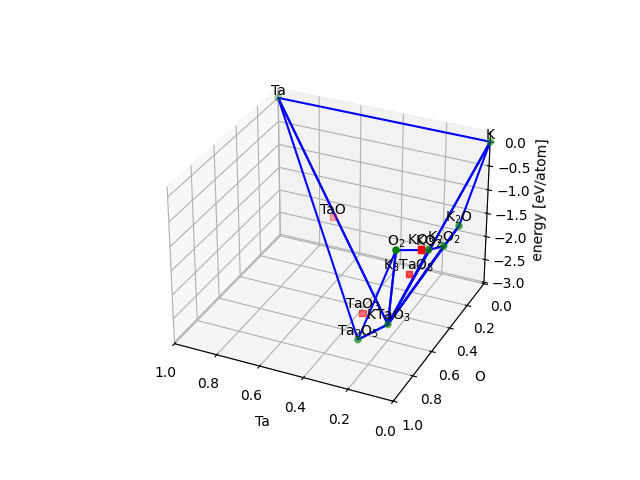
Phase diagrams#
Brillouin zones#
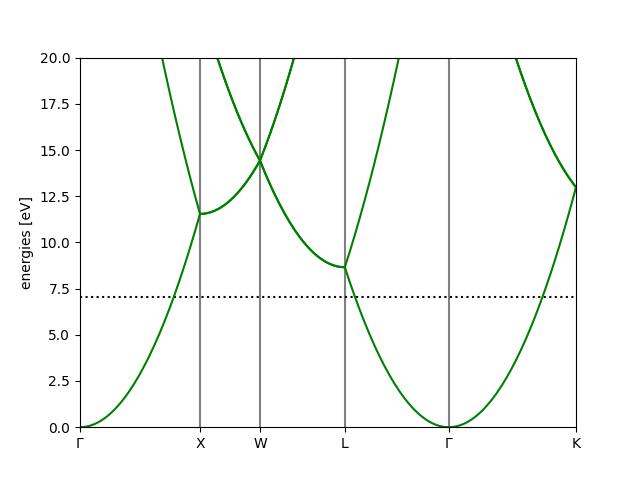
Band-structure#
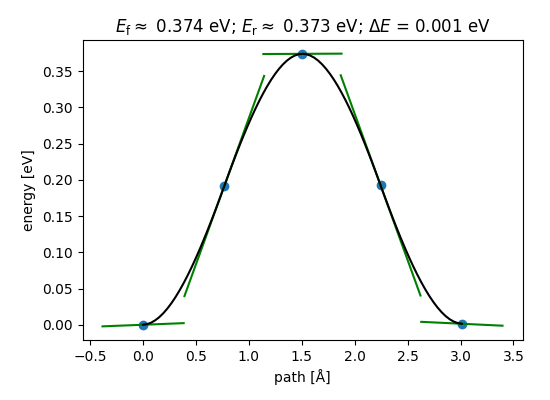
Nudged elastic band calculations#
Nanoparticle#
Pretty pictures#
Periodic table#
Fine tuning POV-Ray settings for high quality images#